VPSC is a crystal plasticity code originally written by Ricardo Lebensohn and Carlos Tome from Los Alamos National Laboratory - USA.
Original code can be requested to lebenso@lanl.gov
https://public.lanl.gov/lebenso/
Import the orientations generated by VPSC
Running a simulation in VPSC usually results in an output file TEX_PH1.OUT which contains multiple sets of orientations for different strain levels. As these files does not contain any information on the crystal symmetry we need to specify it first
cs = crystalSymmetry('222', [4.762 10.225 5.994],'mineral', 'olivine');In the next step the orientations are imported and converted into a list of ODFs using the command ODF.load.
% put in here the path to the VPSC output files
path2file = [mtexDataPath filesep 'VPSC'];
odf = SO3Fun.load([path2file filesep 'TEX_PH1.OUT'],'halfwidth',10*degree,'CS',cs)odf =
1×9 cell array
Columns 1 through 3
{1×1 SO3FunHarmonic} {1×1 SO3FunHarmonic} {1×1 SO3FunHarmonic}
Columns 4 through 6
{1×1 SO3FunHarmonic} {1×1 SO3FunHarmonic} {1×1 SO3FunHarmonic}
Columns 7 through 9
{1×1 SO3FunHarmonic} {1×1 SO3FunHarmonic} {1×1 SO3FunHarmonic}The individual ODFs can be accessed by odf{id}
% lets plot the second ODF
plotSection(odf{2},'sigma','figSize','normal')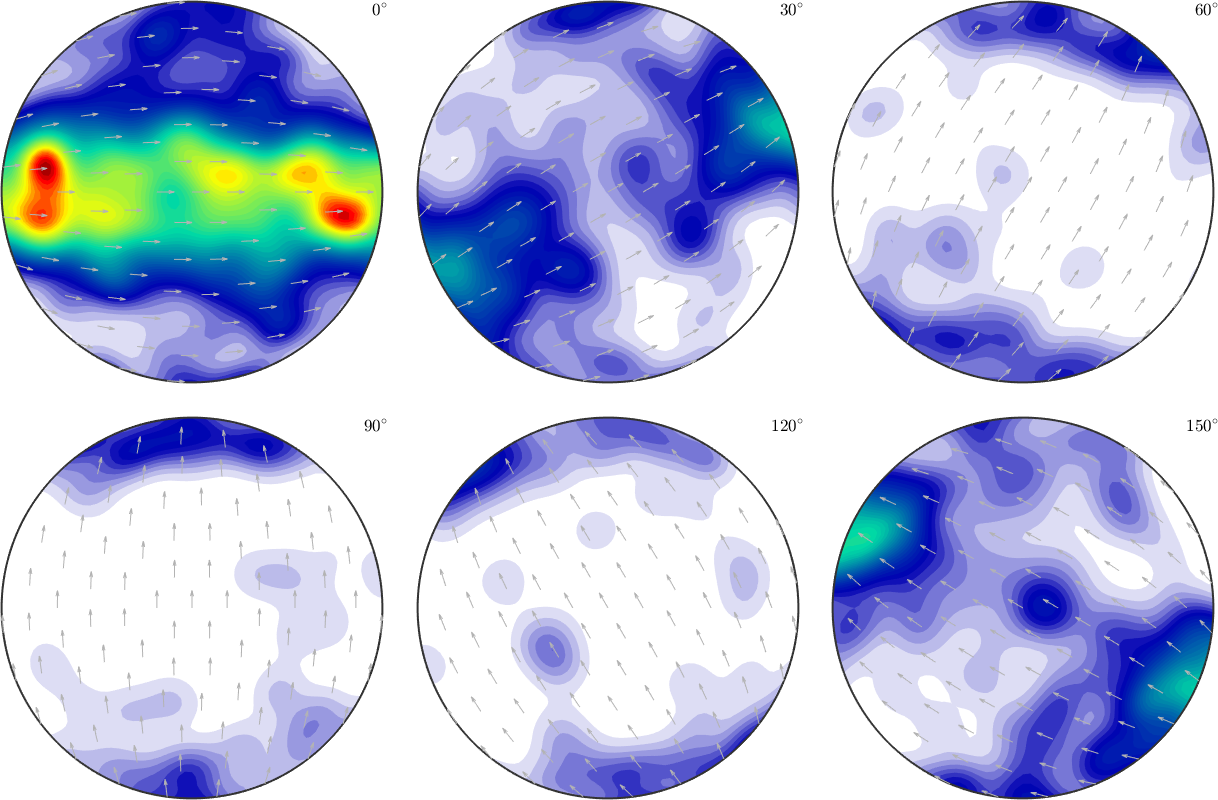
The information about the strain are stored as additional properties within each ODF variable
odf{1}.optans =
struct with fields:
strain: 0.2500
strainEllipsoid: [1.1230 1.1270 0.7500]
strainEllipsoidAngles: [-180 90 -180]
orientations: [1000×1 orientation]
data: [1000×3 double]Compare pole figures during deformation
Next we examine the evaluation of the ODF during the deformation by plotting strain depended pole figures.
% define some crystal directions
h = Miller({1,0,0},{0,1,0},{0,0,1},cs,'uvw');
% generate some figure
fig = newMtexFigure('layout',[4,3],'figSize','huge');
subSet = 1:4;
% plot pole figures for different strain steps
for n = subSet
nextAxis
plotPDF(odf{n},h,'lower','contourf','doNotDraw');
ylabel(fig.children(end-2),['\epsilon = ',xnum2str(odf{n}.opt.strain)]);
end
setColorRange('equal')
mtexColorbar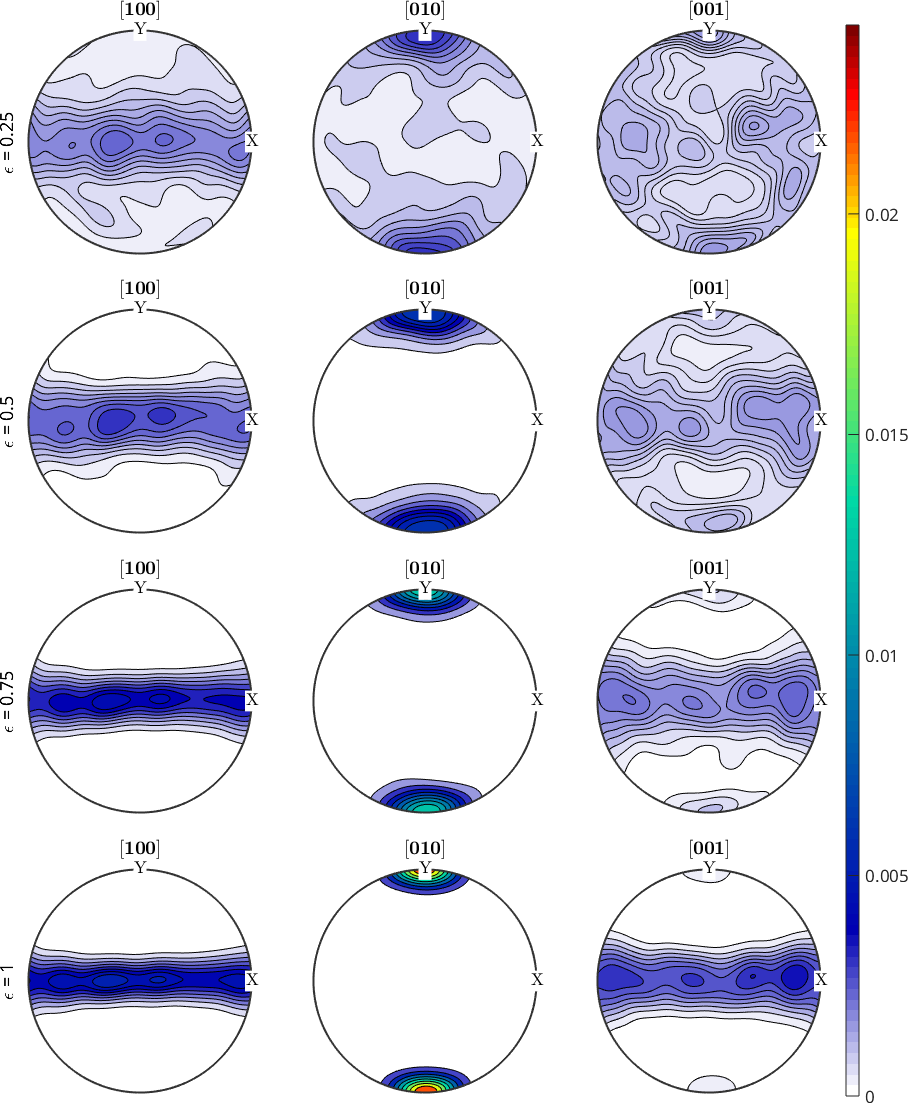
Visualize slip system activity
Alongside with the orientation data VPSC also outputs a file ACT_PH1.OUT which contains the activity of the different slip systems during the deformation. Lets read this file as a table
ACT = readtable([path2file filesep 'ACT_PH1.OUT'],'FileType','text')ACT =
9×11 table
STRAIN AVACS MODE1 MODE2 MODE3 MODE4 MODE5 MODE6 MODE7 MODE8 MODE9
______ _____ _____ _____ _____ _____ _____ _____ _____ _____ _____
0 2.835 0.337 0.31 0.309 0.011 0.012 0.007 0.003 0.002 0.009
0.25 2.766 0.312 0.23 0.417 0.009 0.01 0.007 0.005 0.003 0.009
0.5 2.835 0.317 0.198 0.445 0.007 0.009 0.007 0.007 0.004 0.006
0.75 2.825 0.31 0.131 0.513 0.005 0.007 0.006 0.015 0.007 0.006
1 2.759 0.312 0.075 0.554 0.003 0.005 0.006 0.028 0.013 0.005
1.25 2.746 0.327 0.053 0.546 0.002 0.004 0.005 0.041 0.02 0.002
1.5 2.736 0.37 0.048 0.521 0.002 0.005 0.005 0.033 0.015 0.002
1.75 2.739 0.394 0.046 0.503 0.002 0.005 0.005 0.031 0.013 0.003
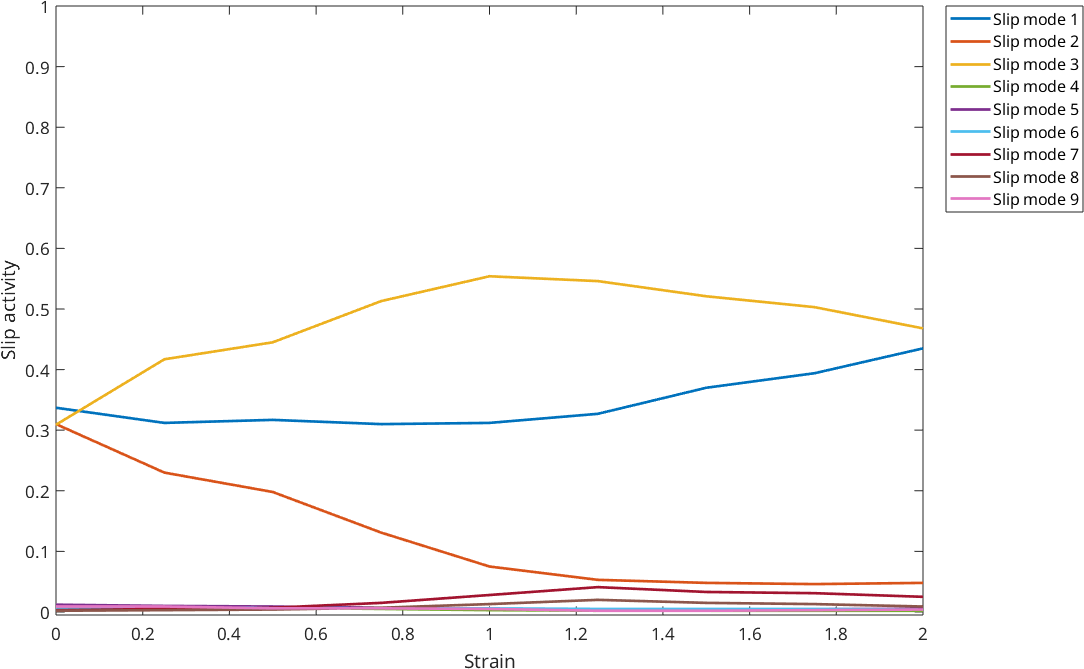
2 2.828 0.435 0.048 0.468 0.002 0.005 0.004 0.025 0.009 0.004and plot the slip activity with respect to the strain for the different modes
% loop though the columns MODE1 ... MOD11
close all
for n = 3: size(ACT,2)
% perform the plotting
plot(ACT.STRAIN, table2array(ACT(:,n)),'linewidth',2,...
'DisplayName',['Slip mode ',num2str(n-2)])
hold on;
end
hold off
% some styling
xlabel('Strain');
ylabel('Slip activity');
legend('show','location','NorthEastOutside');
set(gca,'Ylim',[-0.005 1])
set(gcf,'MenuBar','none','units','normalized','position',[0.25 0.25 0.5 0.5]);
%for only one mode plot, e.g.,mode 3: cs = csapi(STRAIN,MODE{3});fnplt(cs,3,'color','b');hold off;