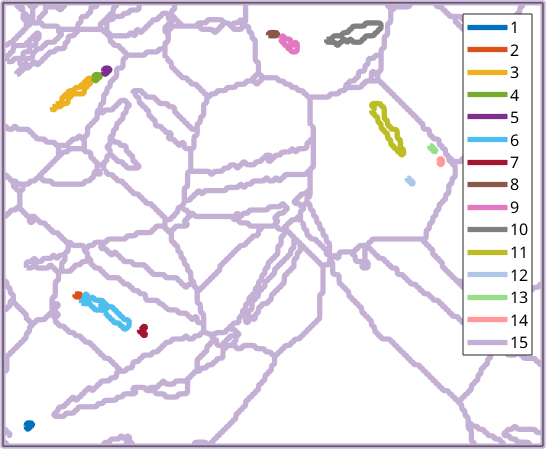
In this section we discus geometric properties that can be derived from grain boundaries. Lets start by importing some EBSD data and computing grain boundaries.
% load some example data
mtexdata twins silent
ebsd.prop = rmfield(ebsd.prop,{'error','bands'});
% detect grains
[grains,ebsd.grainId,ebsd.mis2mean] = calcGrains(ebsd('indexed'));
% smooth them
grains = grains.smooth;
% visualize the grains
plot(grains,grains.meanOrientation)
% extract all grain boundaries
gB = grains.boundary;
hold on
plot(gB,'LineWidth',2)
hold off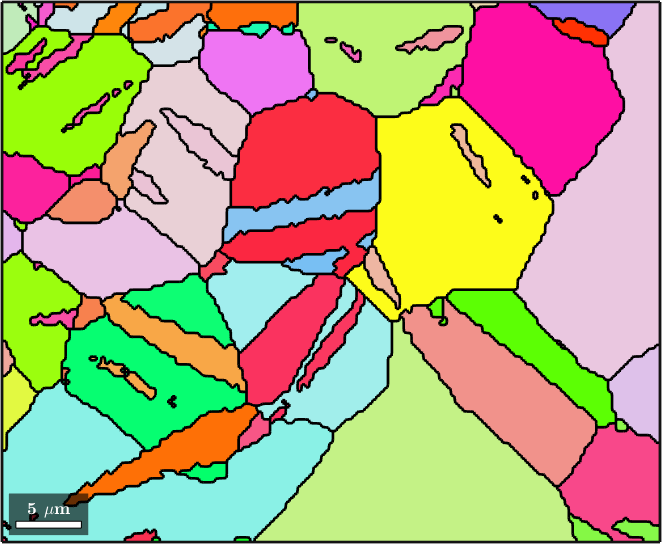
Property overview
A variable of type grainBoundary contains the following properties
|
ebsdId |
neighboring pixel ids |
phaseId |
neighboring phase ids |
|
grainId |
neighboring grain ids |
F |
vertices ids of the segments |
|
length of each segment |
direction |
direction of each segment |
|
|
midPoint |
mid point of the segment |
curvature of each segment |
|
|
misorientation |
between ebsdId(:,1) and ebsdId(:,2) |
||
|
componentId |
connected component id |
componentSize |
connected component size |
The first three properties refer to \(N \times 2\) matrices where \(N\) is the number of boundary segments. Each row of these matrices contains the information about the EBSD data, and grain data on both sides of the grain boundary. To illustrate this consider the grain boundary of one specific grain
gB8 = grains(8).boundarygB8 = grainBoundary
Segments length mineral 1 mineral 2
8 2 µm Magnesium MagnesiumThis boundary consists of 6 segments and hence ebsdId forms a 8x2 matrix
gB8.ebsdIdans =
1010 1177
1010 1009
843 842
843 676
843 844
1011 844
1011 1012
1011 1178It is important to understand that the id is not necessarily the same as the index in the list. In order to index an variable of type EBSD by id and not by index the following syntax has to be used
ebsd('id',gB8.ebsdId)ans = EBSD (y↑→x)
Phase Orientations Mineral Color Symmetry Crystal reference frame
1 16 (100%) Magnesium LightSkyBlue 6/mmm X||a*, Y||b, Z||c*
Id Phase orientation bc bs mad grainId mis2mean
1010 1177 1 1 (113.7°,15.5°,219.3°) 164 168 158 171 0.5 0.3 8 21 (217.3°,0.2°,142.9°)
1010 1009 1 1 (113.7°,15.5°,219.3°) 164 156 158 160 0.5 0.4 8 21 (217.3°,0.2°,142.9°)
843 842 1 1 (115.3°,15.6°,218.2°) 170 167 176 170 0.7 0.5 8 21 (55.9°,0.2°,303.8°)
843 676 1 1 (115.3°,15.6°,218.2°) 170 176 176 196 0.7 0.5 8 21 (55.9°,0.2°,303.8°)
843 844 1 1 (115.3°,15.6°,218.2°) 170 174 176 197 0.7 0.3 8 21 (55.9°,0.2°,303.8°)
1011 844 1 1 (114.7°,15.7°,218.5°) 182 174 174 197 0.5 0.3 8 21 (339.9°,0.1°,20.2°)
1011 1012 1 1 (114.7°,15.7°,218.5°) 182 176 174 168 0.5 0.4 8 21 (339.9°,0.1°,20.2°)
1011 1178 1 1 (114.7°,15.7°,218.5°) 182 174 174 181 0.5 0.5 8 21 (339.9°,0.1°,20.2°)
Scan unit : um
X x Y x Z : [1.8, 2.7] x [1.2, 2.1] x [0, 0]
Normal vector: (0,0,1)
Square grid :8 x 2Similarly
gB8.grainIdans =
8 21
8 21
8 21
8 21
8 21
8 21
8 21
8 21results in 9x2 matrix indicating that grain 8 is an inclusion of grain 21.
plot(grains(8),'FaceColor','DarkBlue','micronbar','off')
hold on
plot(grains(21),'FaceColor','LightCoral')
hold off
Grain boundary misorientations
The grain boundary misorientation defined as the misorientation between the orientations corresponding to ids in first and second column of ebsdId, i.e. following two commands should give the same result
gB8(1).misorientation
inv(ebsd('id',gB8.ebsdId(1,2)).orientations) .* ebsd('id',gB8.ebsdId(1,1)).orientationsans = misorientation (Magnesium → Magnesium)
antipodal: true
Bunge Euler angles in degree
phi1 Phi phi2
330.069 86.0994 150.22
ans = misorientation (Magnesium → Magnesium)
Bunge Euler angles in degree
phi1 Phi phi2
330.069 86.0994 150.22Note that in the first result the antipodal flag is true while it is false in the second result.
Obviously, misorientations of a list of grain boundaries can only be extracted if all of them have the same type of phase transition. Let us consider only Magnesium to Magnesium grain boundaries, i.e., ommit all grain boundaries to an not indexed region.
gB_Mg = gB('Magnesium','Magnesium')gB_Mg = grainBoundary
Segments length mineral 1 mineral 2
3219 837 µm Magnesium MagnesiumThen the misorientation angles can be plotted by
plot(gB_Mg,gB_Mg.misorientation.angle./degree,'linewidth',4)
mtexColorbar('title','misorientation angle (°)')
Geometric properties
The direction property of the boundary segments is useful when checking for tilt and twist boundaries, i.e., when we want to compare the misorientation axis with the interface between the grains
% compute misorientation axes in specimen coordinates
ori = ebsd('id',gB_Mg.ebsdId).orientations;
axes = axis(ori(:,1),ori(:,2),'antipodal')
% plot the angle between the misorientation axis and the boundary direction
plot(gB_Mg,angle(gB_Mg.direction,axes),'linewidth',4)axes = vector3d (y↑→x)
size: 3219 x 1
antipodal: true
We observe that the angle is quite oscillatory. This is because of the stair casing effect when reconstructing grains from gridded EBSD data. The weaken this effect we may average the segment directions using the command calcMeanDirection
% plot the angle between the misorientation axis and the boundary direction
plot(gB_Mg,angle(gB_Mg.calcMeanDirection(4),axes),'linewidth',4)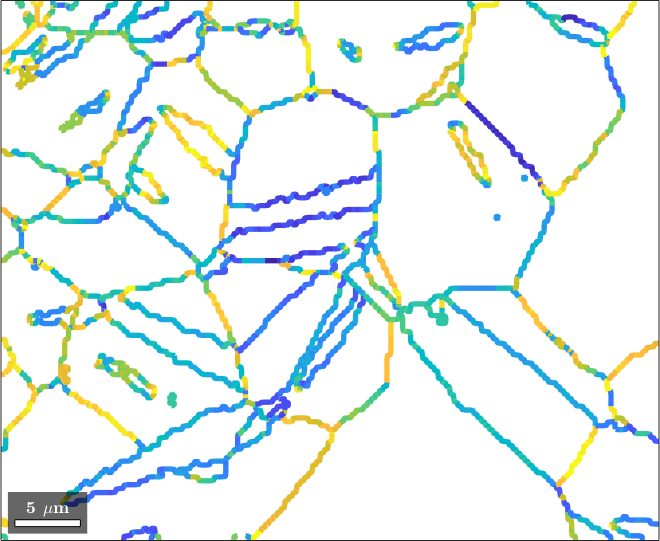
The midPoint property is useful when TODO:
While the command length(gB_Mg) gives the total number of all Magnesium to Magnesium grain boundary segments the command segLength(gB_Mg) gives the length of each segment in µm. The total length of all Magnesium to Magnesium grain boundary segments is hence
sum(gB_Mg.segLength)ans =
837.4744Connected components
TODO: explain this in more detail
components = unique(gB.componentId);
for cId = components.'
plot(gB(gB.componentId == cId),'lineColor',ind2color(cId),...
'micronbar','off','lineWidth',4,'displayName',num2str(cId))
hold on
end
hold off