Schmid Factor Analysis
This script describes how to analyze Schmid factors.
| On this page ... |
| Geometric Definition |
| Stress Tensor |
| Active Slip System |
| The Schmid factor for EBSD data |
| Strain based analysis on the same data set |
Geometric Definition
Let us assume a Nickel crystal
CS = crystalSymmetry('cubic',[3.523,3.523,3.523],'mineral','Nickel')
CS = crystalSymmetry mineral : Nickel symmetry: m-3m a, b, c : 3.5, 3.5, 3.5
Since Nickel is fcc a dominant slip system is given by the slip plane normal
n = Miller(1,1,1,CS,'hkl')n = Miller size: 1 x 1 mineral: Nickel (m-3m) h 1 k 1 l 1
and the slip direction (which needs to be orthogonal)
d = Miller(0,-1,1,CS,'uvw')d = Miller size: 1 x 1 mineral: Nickel (m-3m) u 0 v -1 w 1
For tension in direction 123
r = normalize(vector3d(1,2,3))
r = vector3d
size: 1 x 1
x y z
0.267261 0.534522 0.801784
the Schmid factor for the slip system [0-11](111) is defined by
tau = dot(d,r,'noSymmetry') * dot(n,r,'noSymmetry')
tau =
0.4286
The same computation can be performed by defining the slip system as an MTEX variable
sS = slipSystem(d,n)
sS = slipSystem mineral: Nickel (m-3m) size: 1 x 1 u v w | h k l CRSS 0 -1 1 1 1 1 1
and using the command SchmidFactor
sS.SchmidFactor(r)
ans =
0.1750
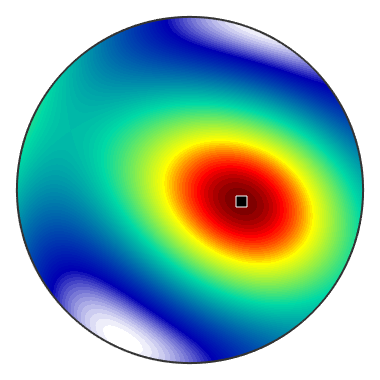
Ommiting the tension direction r the command SchmidFactor returns the Schmid factor as a spherical function
SF = sS.SchmidFactor % plot the Schmid factor in dependency of the tension direction plot(SF) % find the tension directions with the maximum Schmid factor [SFMax,pos] = max(SF) % and annotate them annotate(pos)
SF = S2FunHarmonic
bandwidth: 64
antipodal: true
SFMax =
0.5000
pos = vector3d
size: 1 x 1
antipodal: true
x y z
0.408421 -0.0919374 0.908152
Stress Tensor
Instead by the tension direction the stress might be specified by a stress tensor
sigma = stressTensor.uniaxial(vector3d.Z)
sigma = stressTensor rank: 2 (3 x 3) 0 0 0 0 0 0 0 0 1
Then the Schmid factor for the slip system sS and the stress tensor sigma is computed by
sS.SchmidFactor(sigma)
ans =
0.4082
Active Slip System
In general a crystal contains not only one slip system but at least all symmetrically equivalent ones. Those can be computed with
sSAll = sS.symmetrise('antipodal')sSAll = slipSystem mineral: Nickel (m-3m) size: 12 x 1 u v w | h k l CRSS 0 -1 1 1 1 1 1 1 0 -1 1 1 1 1 -1 1 0 1 1 1 1 -1 1 0 1 1 -1 1 -1 0 -1 1 1 -1 1 0 -1 -1 1 1 -1 1 0 -1 1 -1 1 1 1 -1 0 -1 -1 1 1 1 -1 -1 0 -1 1 1 1 1 0 -1 1 -1 1 1 -1 -1 0 1 -1 1 1 0 -1 -1 1 -1 1 1
The option antipodal indicates that Burgers vectors in oposite direction should not be distinguished. Now
tau = sSAll.SchmidFactor(r)
tau =
Columns 1 through 7
0.1750 -0.3499 0.1750 0.0000 -0.0000 -0.0000 0.1166
Columns 8 through 12
-0.4666 -0.3499 -0.1166 -0.1750 -0.2916
returns a list of Schmid factors and we can find the slip system with the largest Schmid factor using
[tauMax,id] = max(abs(tau)) sSAll(id)
tauMax =
0.4666
id =
8
ans = slipSystem
mineral: Nickel (m-3m)
size: 1 x 1
u v w | h k l CRSS
-1 0 -1 -1 1 1 1
The above computation can be easily extended to a list of tension directions
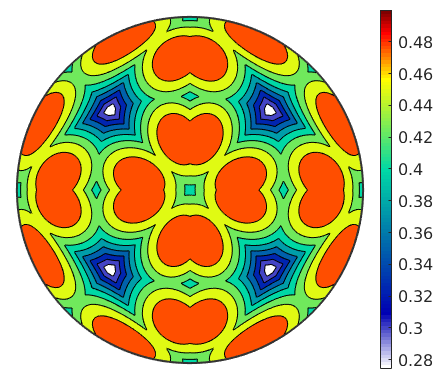
% define a grid of tension directions r = plotS2Grid('resolution',0.5*degree,'upper'); % compute the Schmid factors for all slip systems and all tension % directions tau = sSAll.SchmidFactor(r); % tau is a matrix with columns representing the Schmid factors for the % different slip systems. Lets take the maximum rhowise [tauMax,id] = max(abs(tau),[],2); % vizualize the maximum Schmid factor contourf(r,tauMax) mtexColorbar
We may also plot the index of the active slip system
pcolor(r,id)
mtexColorMap black2white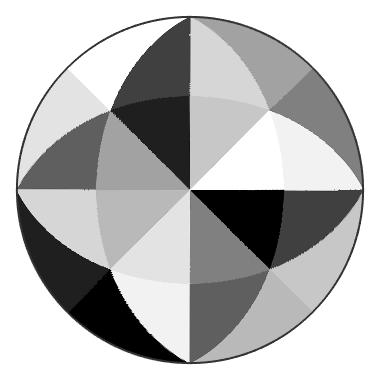
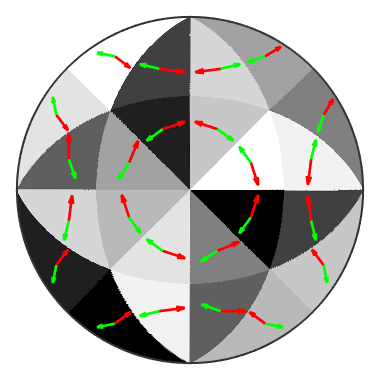
and observe that within the fundamental sectors the active slip system remains the same. We can even visualize the the plane normal and the slip direction
% if we ommit the option antipodal we can distinguish % between the oposite burger vectors sSAll = sS.symmetrise % take as directions the centers of the fundamental regions r = symmetrise(CS.fundamentalSector.center,CS); % compute the Schmid factor tau = sSAll.SchmidFactor(r); % here we do not need to take the absolut value since we consider both % burger vectors +/- b [~,id] = max(tau,[],2); % plot active slip plane in red hold on quiver(r,sSAll(id).n,'ArrowSize',0.25,'LineWidth',2,'Color','r'); % plot active slip direction in green hold on quiver(r,sSAll(id).b.normalize,'ArrowSize',0.12,'LineWidth',2,'Color','g'); hold off
sSAll = slipSystem mineral: Nickel (m-3m) size: 24 x 1 u v w | h k l CRSS 0 -1 1 1 1 1 1 1 0 -1 1 1 1 1 -1 1 0 1 1 1 1 0 1 -1 1 1 1 1 -1 0 1 1 1 1 1 1 -1 0 1 1 1 1 -1 1 0 1 1 -1 1 -1 0 -1 1 1 -1 1 0 -1 -1 1 1 -1 1 1 0 1 1 1 -1 1 0 1 1 1 1 -1 1 1 -1 0 1 1 -1 1 0 -1 1 -1 1 1 1 -1 0 -1 -1 1 1 1 -1 -1 0 -1 1 1 1 1 0 1 -1 1 1 1 1 1 0 -1 1 1 1 0 1 -1 -1 1 1 1 1 0 -1 1 -1 1 1 -1 -1 0 1 -1 1 1 0 -1 -1 1 -1 1 1 1 1 0 1 -1 1 1 0 1 1 1 -1 1 1 -1 0 1 1 -1 1 1
If we perform this computation in terms of spherical functions we obtain
% ommiting |r| gives us a list of 12 spherical functions tau = sSAll.SchmidFactor % now we take the max of the absolute value over all these functions contourf(max(abs(tau),[],1),'upper') mtexColorbar
tau = S2FunHarmonic size: 24 x 1 bandwidth: 64 antipodal: true
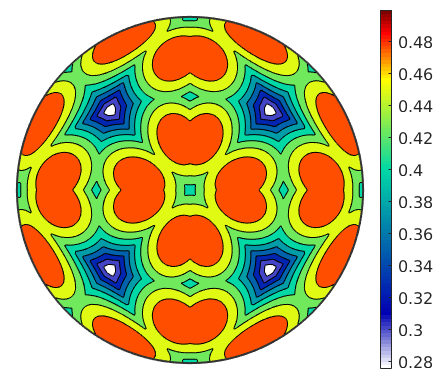
The Schmid factor for EBSD data
So far we have always assumed that the stress tensor is already given relatively to the crystal coordinate system. Next, we want to examine the case where the stress is given in specimen coordinates and we know the orientation of the crystal. Lets import some EBSD data and computet the grains
mtexdata csl % take some subset ebsd = ebsd(ebsd.inpolygon([0,0,200,50])) grains = calcGrains(ebsd); grains = smooth(grains,5); plot(ebsd,ebsd.orientations,'micronbar','off') hold on plot(grains.boundary,'linewidth',2) hold off
ebsd = EBSD
Phase Orientations Mineral Color Symmetry Crystal reference frame
-1 10251 (100%) iron light blue m-3m
Properties: ci, error, iq, x, y
Scan unit : um
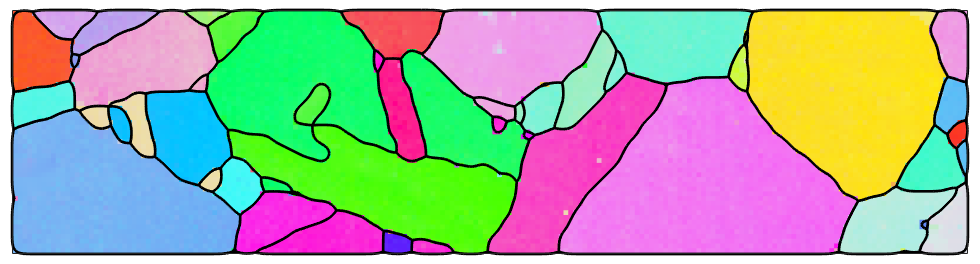
We want to consider the following slip systems
sS = slipSystem.fcc(ebsd.CS) sS = sS.symmetrise;
sS = slipSystem mineral: iron (m-3m) size: 1 x 1 u v w | h k l CRSS 0 1 -1 1 1 1 1
Since, those slip systems are in crystal coordinates but the stress tensor is in specimen coordinates we either have to rotate the slip systems into specimen coordinates or the stress tensor into crystal coordinates. In the following sections we will demonstrate both ways. Lets start with the first one
% rotate slip systems into specimen coordinates
sSLocal = grains.meanOrientation * sSsSLocal = slipSystem CRSS: 1 size: 71 x 24
These slip systems are now arranged in matrix form where the rows corrspond to the crystal reference frames of the different grains and the rows are the symmetrically equivalent slip systems. Computing the Schmid faktor we end up with a matrix of the same size
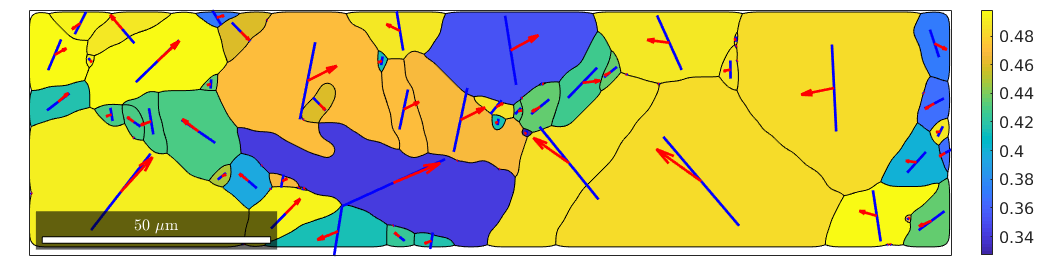
% compute Schmid factor sigma = stressTensor.uniaxial(vector3d.X) SF = sSLocal.SchmidFactor(sigma); % take the maxium allong the rows [SFMax,active] = max(SF,[],2); % plot the maximum Schmid factor plot(grains,SFMax,'micronbar','off','linewidth',2) mtexColorbar location southoutside
sigma = stressTensor rank: 2 (3 x 3) 1 0 0 0 0 0 0 0 0
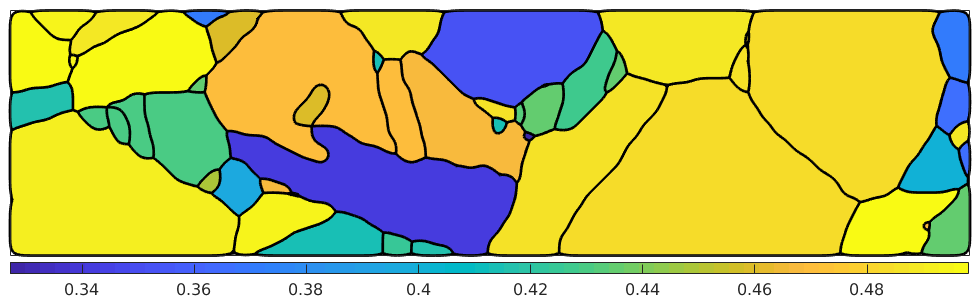
Next we want to visualize the active slip systems.
% take the active slip system and rotate it in specimen coordinates sSactive = grains.meanOrientation .* sS(active); hold on % visualize the trace of the slip plane quiver(grains,sSactive.trace,'color','b') % and the slip direction quiver(grains,sSactive.b,'color','r') hold off
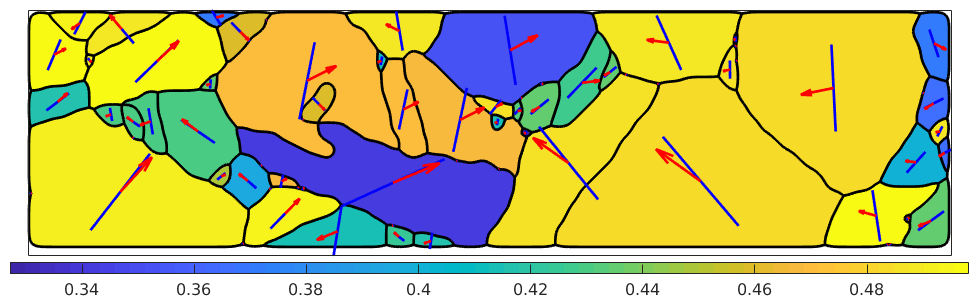
We observe that the Burgers vector is in most case aligned with the trace. In those cases where trace and Burgers vector are not aligned the slip plane is not perpendicular to the surface and the Burgers vector sticks out of the surface.
Next we want to demonstrate the alternative route
% rotate the stress tensor into crystal coordinates
sigmaLocal = inv(grains.meanOrientation) * sigmasigmaLocal = stressTensor size : 71 x 1 rank : 2 (3 x 3) mineral: iron (m-3m)
This becomes a list of stress tensors with respect to crystal coordinates - one for each grain. Now we have both the slip systems as well as the stress tensor in crystal coordiantes and can compute the Schmid factor
% the resulting matrix is the same as above SF = sS.SchmidFactor(sigmaLocal); % and hence we may proceed analogously % take the maxium allong the rows [SFMax,active] = max(SF,[],2); % plot the maximum Schmid factor plot(grains,SFMax) mtexColorbar % take the active slip system and rotate it in specimen coordinates sSactive = grains.meanOrientation .* sS(active); hold on % visualize the trace of the slip plane quiver(grains,sSactive.trace,'color','b') % and the slip direction quiver(grains,sSactive.b,'color','r') hold off
Strain based analysis on the same data set
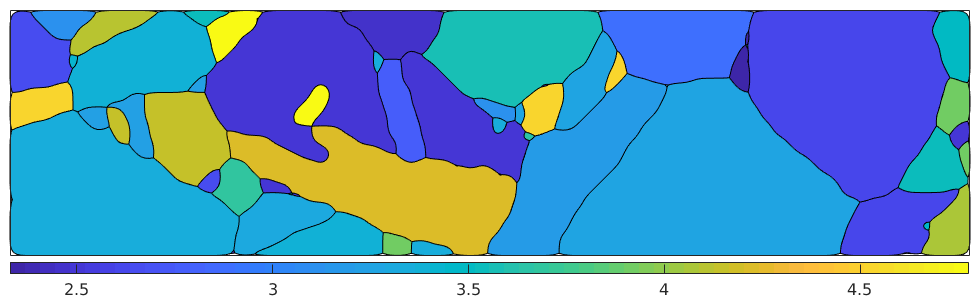
eps = strainTensor(diag([1,0,-1])) epsCrystal = inv(grains.meanOrientation) * eps [M, b, W] = calcTaylor(epsCrystal, sS); plot(grains,M,'micronbar','off') mtexColorbar southoutside
eps = strainTensor rank: 2 (3 x 3) 1 0 0 0 0 0 0 0 -1 epsCrystal = strainTensor size : 71 x 1 rank : 2 (3 x 3) mineral: iron (m-3m) computing Taylor factor: 100%
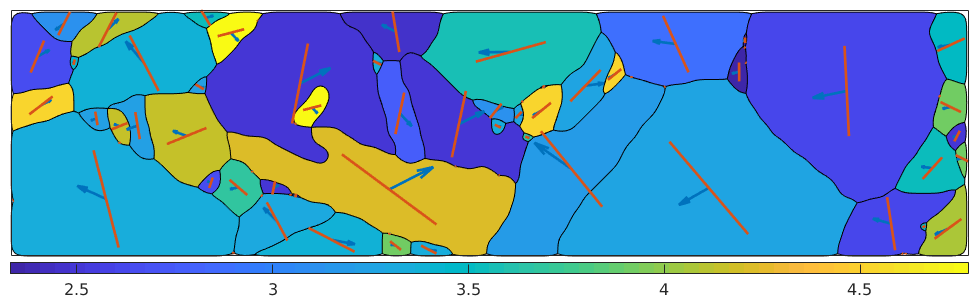
[ bMax , bMaxId ] = max( b , [ ] , 2 ) ; sSGrains = grains.meanOrientation .* sS(bMaxId) ; hold on quiver ( grains , sSGrains.b) quiver ( grains , sSGrains.trace) hold off
| DocHelp 0.1 beta |